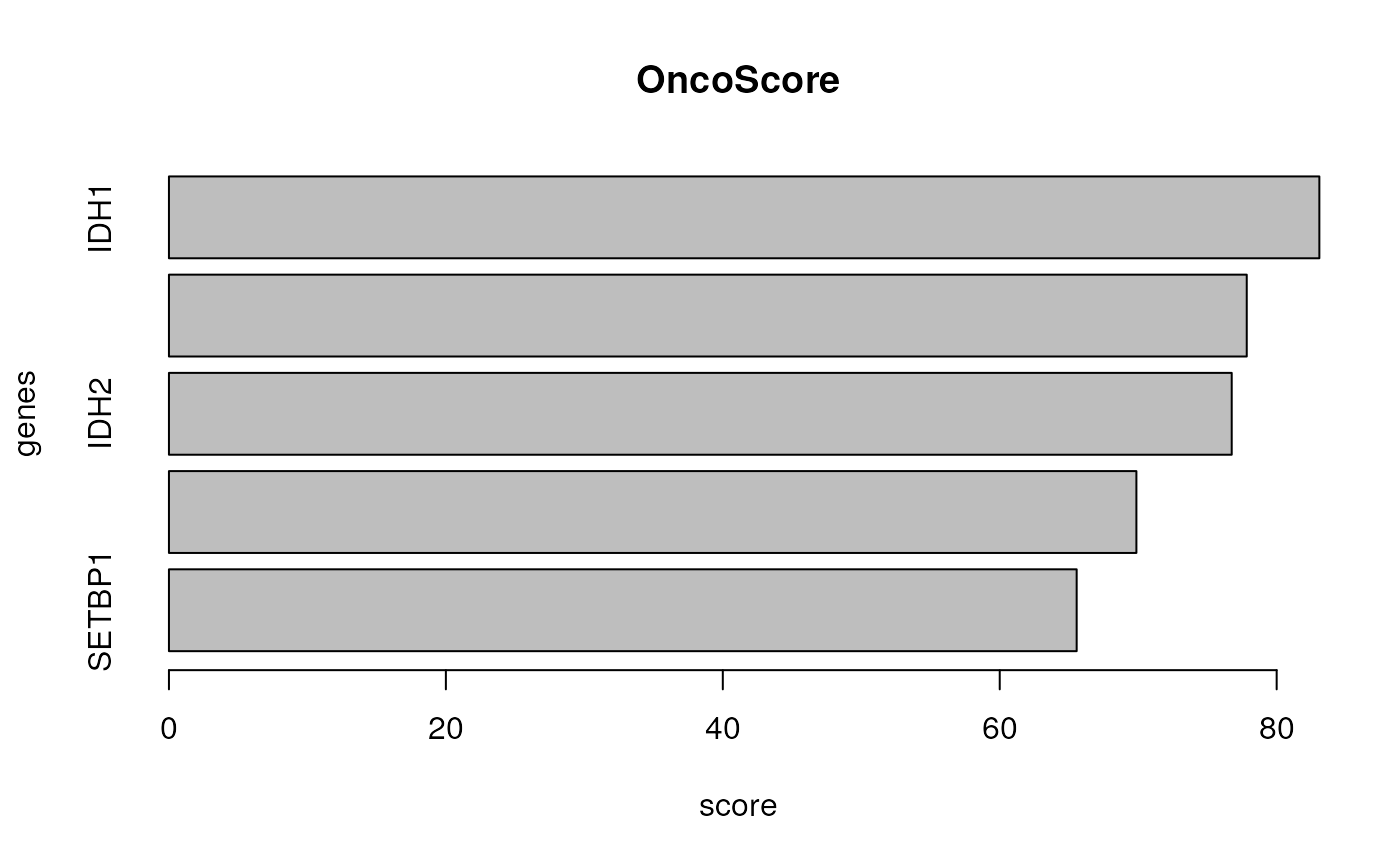
plot the OncoScore for a list of genes
# S3 method for class 'oncoscore'
plot(
x,
gene.number = 5,
main = "OncoScore",
xlab = "score",
ylab = "genes",
file = NA,
...
)Arguments
Value
A plot
Examples
data(query)
result = compute.oncoscore(query)
#> ### Processing data
#> ### Computing frequencies scores
#> ### Estimating oncogenes
#> ### Results:
#> ASXL1 -> 77.8392
#> IDH1 -> 83.08351
#> IDH2 -> 76.75356
#> SETBP1 -> 65.556
#> TET2 -> 69.86954
plot.oncoscore(result)