Running OncoScore
Luca De Sano
Carlo Gambacorti Passerini
Rocco Piazza
Daniele Ramazzotti
Roberta Spinelli
October 15, 2025
Source:vignettes/v2_running_OncoScore.Rmd
v2_running_OncoScore.RmdThe OncoScore analysis consists of two parts. One can estimate a score to asses the oncogenic potential of a set of genes, given the lecterature knowledge, at the time of the analysis, or one can study the trend of such score over time.
We next present the two analysis and we conclude with showing the capabilities of the tool to visualize the results.
First we load the library.
The query that we show next retrieves from PubMed the citations, at the time of the query, for a list of genes in cancer related and in all the documents.
query = perform.query(c("ASXL1","IDH1","IDH2","SETBP1","TET2"))## ### Starting the queries for the selected genes.
##
## ### Performing queries for cancer literature
## Number of papers found in PubMed for ASXL1 was: 1577
## Number of papers found in PubMed for IDH1 was: 5681
## Number of papers found in PubMed for IDH2 was: 2362
## Number of papers found in PubMed for SETBP1 was: 303
## Number of papers found in PubMed for TET2 was: 2716
##
## ### Performing queries for all the literature
## Number of papers found in PubMed for ASXL1 was: 1757
## Number of papers found in PubMed for IDH1 was: 6058
## Number of papers found in PubMed for IDH2 was: 2697
## Number of papers found in PubMed for SETBP1 was: 399
## Number of papers found in PubMed for TET2 was: 3627OncoScore provides a function to merge gene names if requested by the user. This function is useful when there are aliases in the gene list.
combine.query.results(query, c('IDH1', 'IDH2'), 'new_gene')## CitationsGene CitationsGeneInCancer
## ASXL1 1757 1577
## SETBP1 399 303
## TET2 3627 2716
## new_gene 8755 8043OncoScore also provides a function to retireve the names of the genes in a given portion of a chromosome that can be exploited if we are dealing, e.g., with copy number alterations hitting regions rather than specific genes.
chr13 = get.genes.from.biomart(chromosome=13,start=54700000,end=72800000)Furthermore, one can also automatically perform the OncoScore analysis on chromosomic regions as follows:
result = compute.oncoscore.from.region(10, 100000, 500000)We now compute a score for each of the genes, to estimate their oncogenic potential.
result = compute.oncoscore(query)## ### Processing data
## ### Computing frequencies scores
## ### Estimating oncogenes
## ### Results:
## ASXL1 -> 81.42832
## IDH1 -> 86.31327
## IDH2 -> 79.89451
## SETBP1 -> 67.15076
## TET2 -> 68.55The query that we show next retrieves from PubMed the citations, at specified time points, for a list of genes in cancer related and in all the documents.
query.timepoints = perform.query.timeseries(c("ASXL1","IDH1","IDH2","SETBP1","TET2"),
c("2012/03/01", "2013/03/01", "2014/03/01", "2015/03/01", "2016/03/01"))## ### Starting the queries for the selected genes.
## ### Quering PubMed for timepoint 2012/03/01
## ### Performing queries for cancer literature
## Number of papers found in PubMed for ASXL1 was: 87
## Number of papers found in PubMed for IDH1 was: 414
## Number of papers found in PubMed for IDH2 was: 213
## Number of papers found in PubMed for SETBP1 was: 6
## Number of papers found in PubMed for TET2 was: 173
## ### Performing queries for all the literature
## Number of papers found in PubMed for ASXL1 was: 93
## Number of papers found in PubMed for IDH1 was: 543
## Number of papers found in PubMed for IDH2 was: 308
## Number of papers found in PubMed for SETBP1 was: 12
## Number of papers found in PubMed for TET2 was: 201
## ### Quering PubMed for timepoint 2013/03/01
## ### Performing queries for cancer literature
## Number of papers found in PubMed for ASXL1 was: 136
## Number of papers found in PubMed for IDH1 was: 669
## Number of papers found in PubMed for IDH2 was: 339
## Number of papers found in PubMed for SETBP1 was: 12
## Number of papers found in PubMed for TET2 was: 258
## ### Performing queries for all the literature
## Number of papers found in PubMed for ASXL1 was: 151
## Number of papers found in PubMed for IDH1 was: 808
## Number of papers found in PubMed for IDH2 was: 441
## Number of papers found in PubMed for SETBP1 was: 20
## Number of papers found in PubMed for TET2 was: 307
## ### Quering PubMed for timepoint 2014/03/01
## ### Performing queries for cancer literature
## Number of papers found in PubMed for ASXL1 was: 190
## Number of papers found in PubMed for IDH1 was: 913
## Number of papers found in PubMed for IDH2 was: 455
## Number of papers found in PubMed for SETBP1 was: 30
## Number of papers found in PubMed for TET2 was: 349
## ### Performing queries for all the literature
## Number of papers found in PubMed for ASXL1 was: 211
## Number of papers found in PubMed for IDH1 was: 1062
## Number of papers found in PubMed for IDH2 was: 566
## Number of papers found in PubMed for SETBP1 was: 38
## Number of papers found in PubMed for TET2 was: 436
## ### Quering PubMed for timepoint 2015/03/01
## ### Performing queries for cancer literature
## Number of papers found in PubMed for ASXL1 was: 259
## Number of papers found in PubMed for IDH1 was: 1217
## Number of papers found in PubMed for IDH2 was: 583
## Number of papers found in PubMed for SETBP1 was: 52
## Number of papers found in PubMed for TET2 was: 465
## ### Performing queries for all the literature
## Number of papers found in PubMed for ASXL1 was: 288
## Number of papers found in PubMed for IDH1 was: 1372
## Number of papers found in PubMed for IDH2 was: 702
## Number of papers found in PubMed for SETBP1 was: 68
## Number of papers found in PubMed for TET2 was: 588
## ### Quering PubMed for timepoint 2016/03/01
## ### Performing queries for cancer literature
## Number of papers found in PubMed for ASXL1 was: 325
## Number of papers found in PubMed for IDH1 was: 1536
## Number of papers found in PubMed for IDH2 was: 720
## Number of papers found in PubMed for SETBP1 was: 69
## Number of papers found in PubMed for TET2 was: 592
## ### Performing queries for all the literature
## Number of papers found in PubMed for ASXL1 was: 361
## Number of papers found in PubMed for IDH1 was: 1705
## Number of papers found in PubMed for IDH2 was: 847## Warning in file(file, "r"): URL
## 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=pubmed&term=SETBP1[All+Fields](0001/01/01[PDAT]+:+2016/03/01[PDAT])&retmax=3000000&rettype=FASTA&tool=OncoScore':
## status was 'Failure when receiving data from the peer'## Number of papers found in PubMed for SETBP1 was: 91
## Number of papers found in PubMed for TET2 was: 751We now compute a score for each of the genes, to estimate their oncogenic potential at specified time points.
result.timeseries = compute.oncoscore.timeseries(query.timepoints)## ### Computing oncoscore for timepoint 2012/03/01
## ### Processing data
## ### Computing frequencies scores
## ### Estimating oncogenes
## ### Results:
## ASXL1 -> 79.24251
## IDH1 -> 67.85072
## IDH2 -> 60.79034
## SETBP1 -> 36.05285
## TET2 -> 74.82026
## ### Computing oncoscore for timepoint 2013/03/01
## ### Processing data
## ### Computing frequencies scores
## ### Estimating oncogenes
## ### Results:
## ASXL1 -> 77.6234
## IDH1 -> 74.22432
## IDH2 -> 68.12016
## SETBP1 -> 46.11731
## TET2 -> 73.86744
## ### Computing oncoscore for timepoint 2014/03/01
## ### Processing data
## ### Computing frequencies scores
## ### Estimating oncogenes
## ### Results:
## ASXL1 -> 78.38488
## IDH1 -> 77.41784
## IDH2 -> 71.59791
## SETBP1 -> 63.90384
## TET2 -> 70.91674
## ### Computing oncoscore for timepoint 2015/03/01
## ### Processing data
## ### Computing frequencies scores
## ### Estimating oncogenes
## ### Results:
## ASXL1 -> 78.92304
## IDH1 -> 80.19158
## IDH2 -> 74.26519
## SETBP1 -> 63.90861
## TET2 -> 70.4855
## ### Computing oncoscore for timepoint 2016/03/01
## ### Processing data
## ### Computing frequencies scores
## ### Estimating oncogenes
## ### Results:
## ASXL1 -> 79.43104
## IDH1 -> 81.69642
## IDH2 -> 76.26603
## SETBP1 -> 64.17289
## TET2 -> 70.57627We next plot the scores measuring the oncogenetic potential of the considered genes as a barplot.
plot.oncoscore(result, col = 'darkblue')
Oncogenetic potential of the considered genes.
We finally plot the trend of the scores over the considered times as absolute and values and as variations.
plot.oncoscore.timeseries(result.timeseries)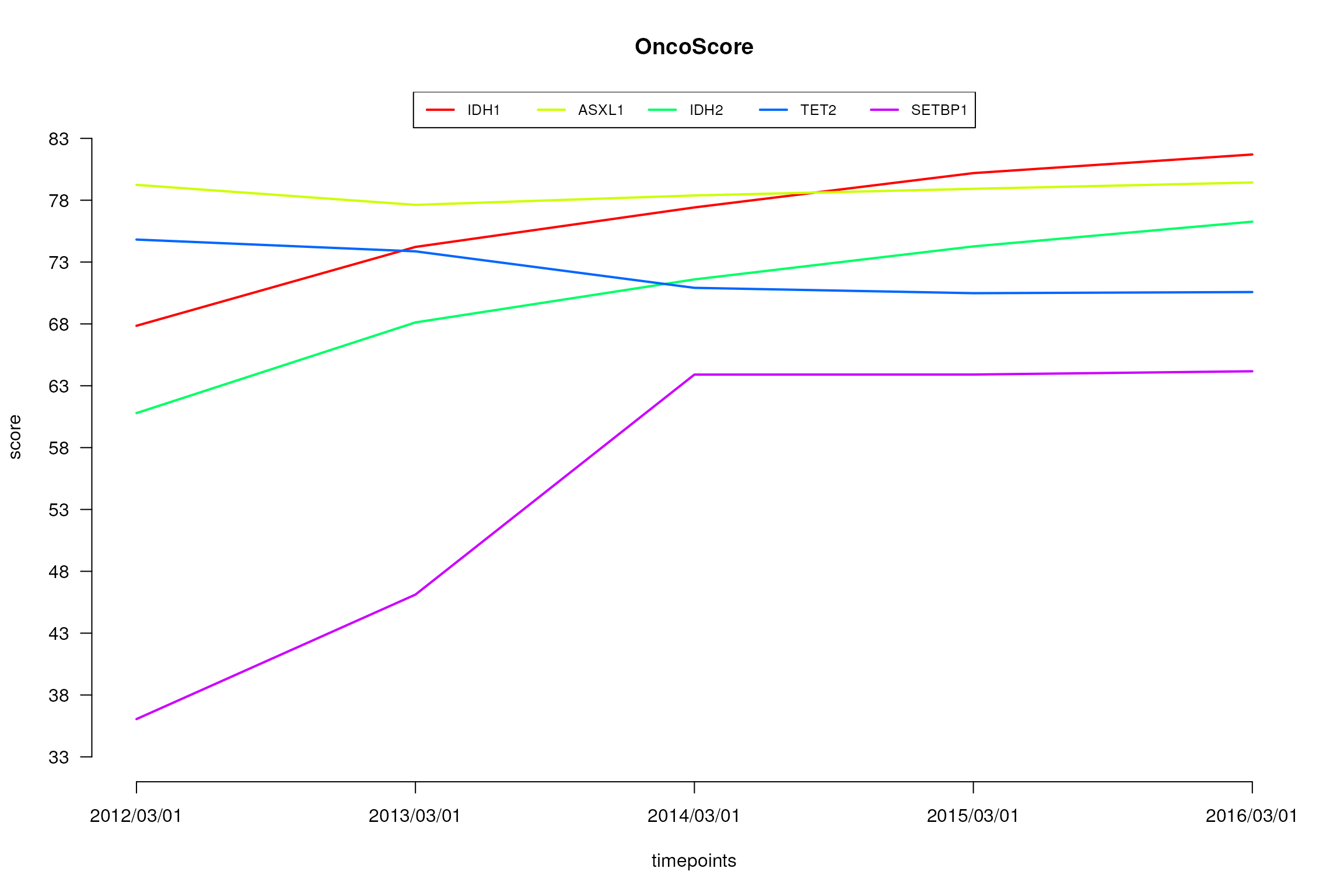
Absolute values of the oncogenetic potential of the considered genes over times.
plot.oncoscore.timeseries(result.timeseries, incremental = TRUE, ylab='absolute variation')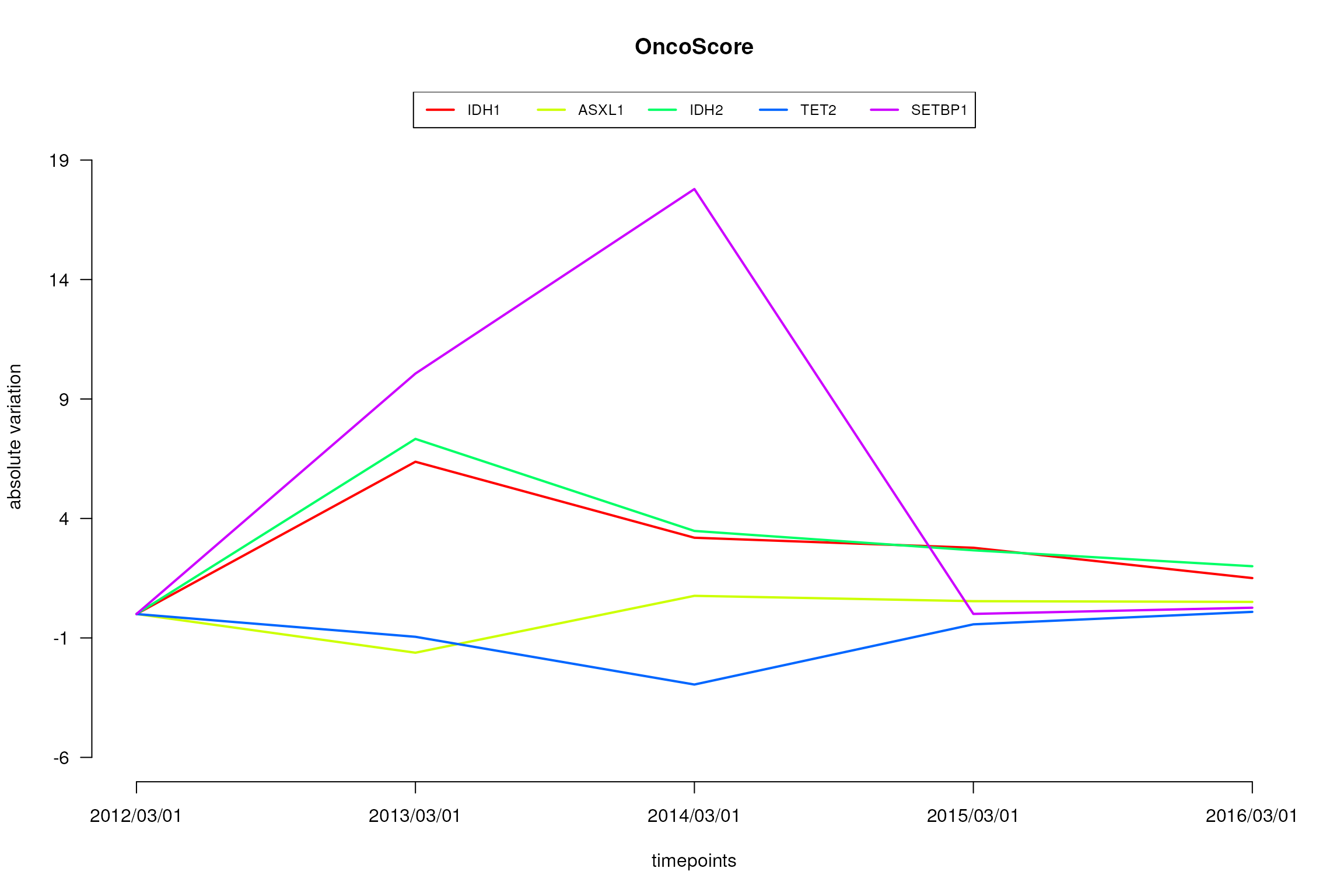
Variations of the oncogenetic potential of the considered genes over times.
plot.oncoscore.timeseries(result.timeseries, incremental = TRUE, relative = TRUE, ylab='relative variation')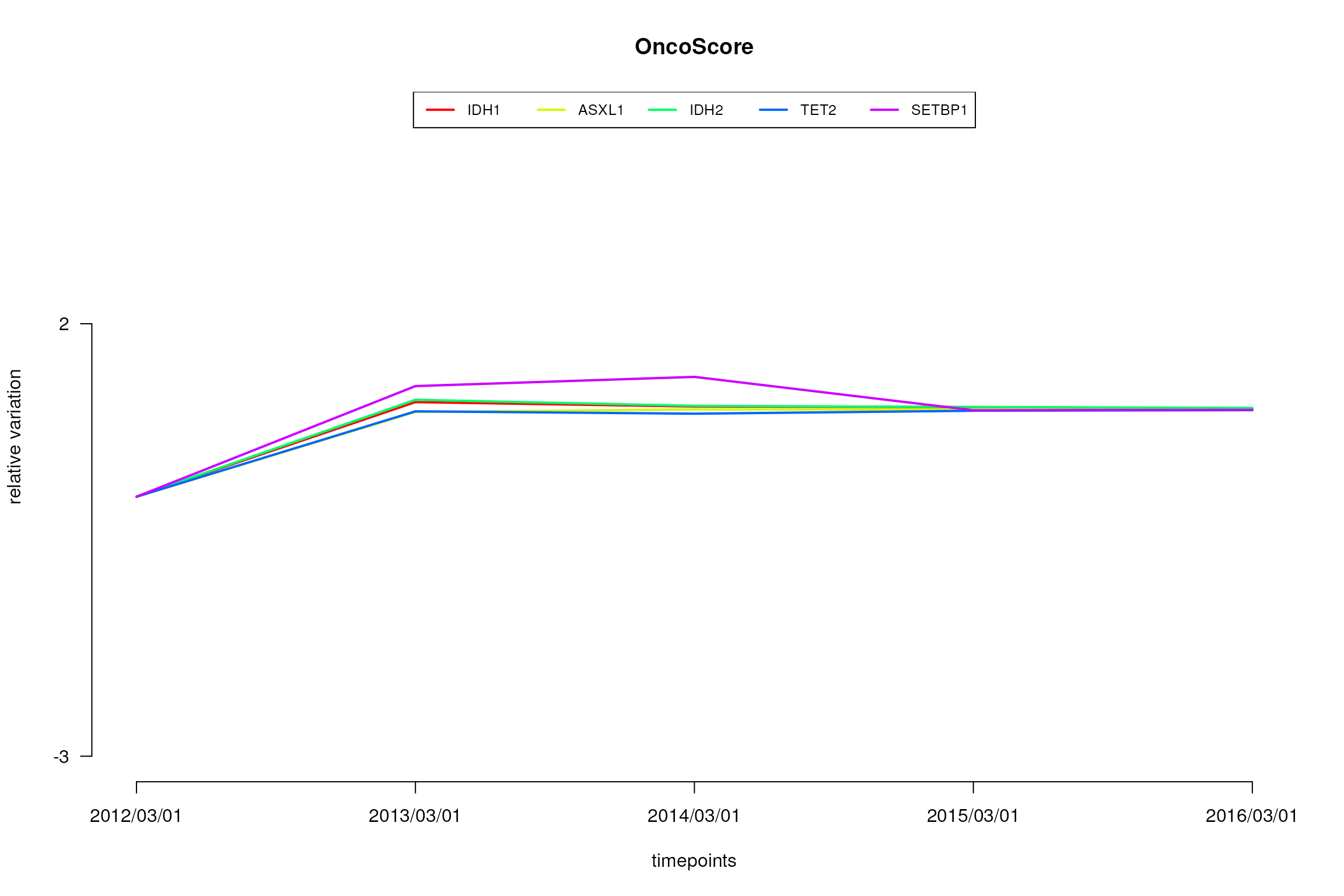
Variations as relative values of the oncogenetic potential of the considered genes over times.